Activatable LNP for Gene Delivery in Extrahepatic Tissues
Miffy Cheng
Investigators
Summary
Current LNP systems have limited capabilities for targeted delivery. Onpattro-like formulations experience ApoE adsorption during systemic circulation, which alters the biodistribution of the genetic payload. This leads to predominantly hepatobiliary clearance, significant uptake in the liver and spleen, and limited deposition in target extrahepatic tissues, making it difficult to reach the desired cell type. Therefore, developing both passive and active targeted delivery strategies would expand the therapeutic applications of LNP-based gene therapy. In our lab, we are expanding our toolbox for achieving specific gene expression through both chemical and molecular biology approaches by introducing light-responsive small molecules within both the carrier and cargo of our formulations. These approaches include lipid prodrugs and ligand-targeting strategies. This research program aims to develop a novel platform technology that increases specificity and reduces toxicity, ultimately broadening the utility of LNP-mRNA against a wide range of diseases.
Influence of Membrane Biophysics of LNP in Cellular Transportation and mRNA Translation
Miffy Cheng
Investigators
Summary
Lipid nanoparticles (LNPs) are vehicles for delivering proteins and nucleic acids. LNPs are made up of building blocks called lipids. Different lipid compositions can affect various biophysical properties of LNPs, such as the membrane fluidity, size and morphologies. When LNPs are administered to biological systems, proteins in the blood bind to and coat LNPs, impacting the functionality and effectiveness of the LNP. Our research aims to identify the proteins that bind onto LNPs, and how that affects the delivery of the genetic cargo into cells of interest. Our lab will tackle this problem by visualizing this interaction using the state-of-the-art imaging methods—such as super-resolution microscopy and cryogenic electron microscopy—and by creating new lipids and techniques to enable effective endosomal escape. This work may provide new insights into RNA biology and nanotechnology, advancing the development of next-generation RNA-based therapeutics.
Bioinspired Nanomaterials to Advance the Oral Delivery of Therapeutic Bacteria
Joel Finbloom
Investigators
Summary
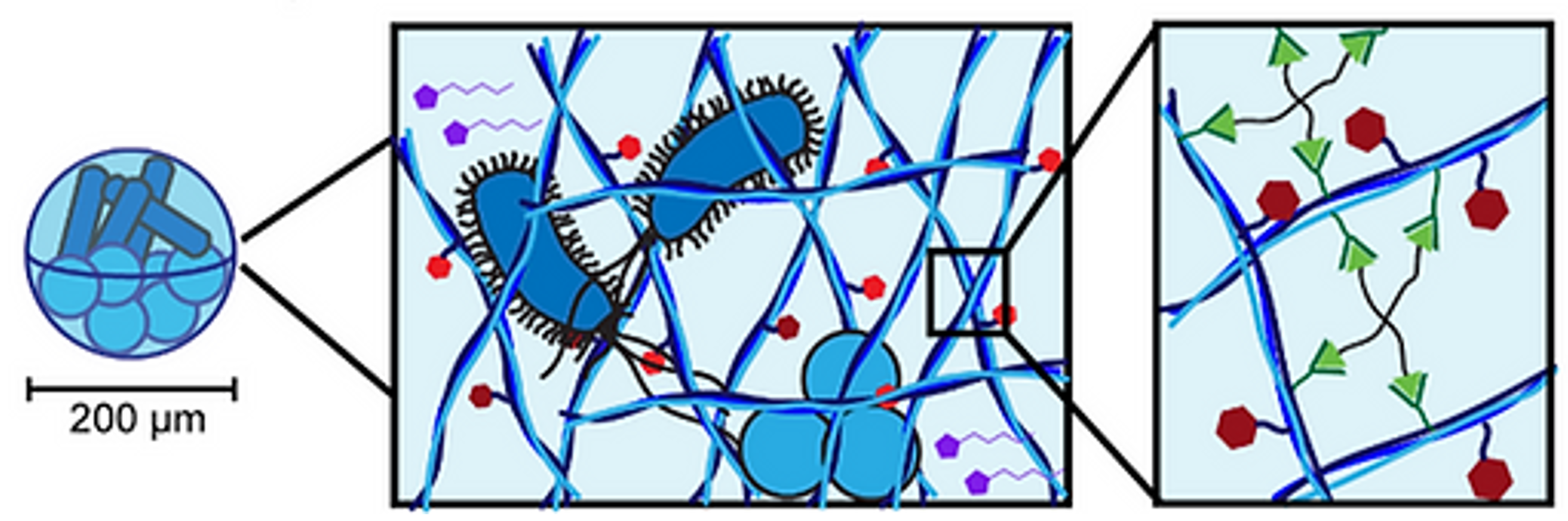
Therapeutic microbes (e.g., probiotics) offer immense promise for the treatment of a variety of diseases, including bacterial infections, autoimmune disorders, and irritable bowel disease. However, the oral delivery of living biotherapeutics such as probiotics is hampered by the immense biological barriers posed by the GI system, such as stomach acid and competition from the native microbiome. We are taking inspiration from natural bacterial communities to design nanostructured microgels with microbe-interfacing functional handles that are capable of encapsulating probiotics to improve their viability, GI retention, and biomedical function.
Improving Antimicrobial Drug Delivery with Microbe-Interfacing Nanomaterials
Joel Finbloom
Investigators
Summary
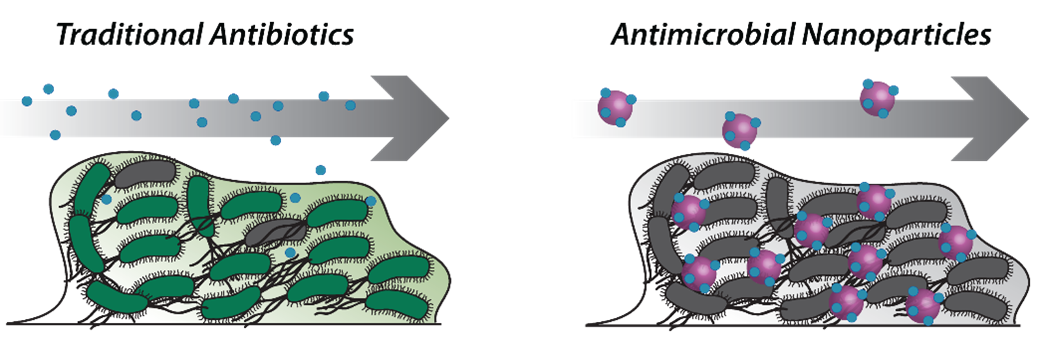
Bacterial biofilm infections are dynamic and heterogeneous bacterial communities, containing a dense extracellular network of polysaccharides, DNA, and proteins. These biofilms are estimated to occur in over 80% of infections and limit antibiotic penetration, leading to 1000x drug resistance. We are designing nanoparticles loaded with antibiotics that interface with bacterial biofilms to direct nanoparticle navigation through the biofilm environment and improve antibiotic delivery efficacies. This approach will aid in our understanding of biofilm microenvironments and develop next generation nanotherapeutics for the targeted treatment of chronic bacterial infections.
Pharmaco-kinetics and -dynamics of radiolabeled biopharmaceuticals measured by SPECT and PET
Urs Häfeli
Investigator
Summary
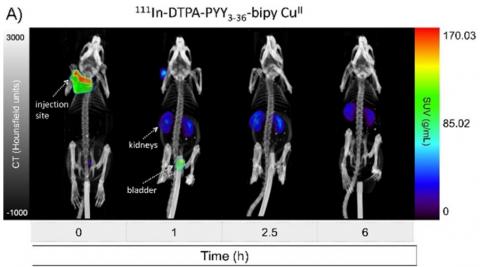
Evaluation of drug exposure (pharmacokinetics, PK) and response (pharmaco-dynamics, PD) is critical to select appropriate molecules, assess safety and efficacy of drug candidates, and design optimal dosing strategies. PK for biopharmaceuticals can be determined by imaging techniques like PET and SPECT, and yields fully quantitative biodistribution and kinetic data. Biopharmaceuticals in this project can include antibodies, peptides, metal-binding small molecules and nanoparticles.
Needle-free Delivery of Proteins
Shyh-Dar Li
Investigator
Summary
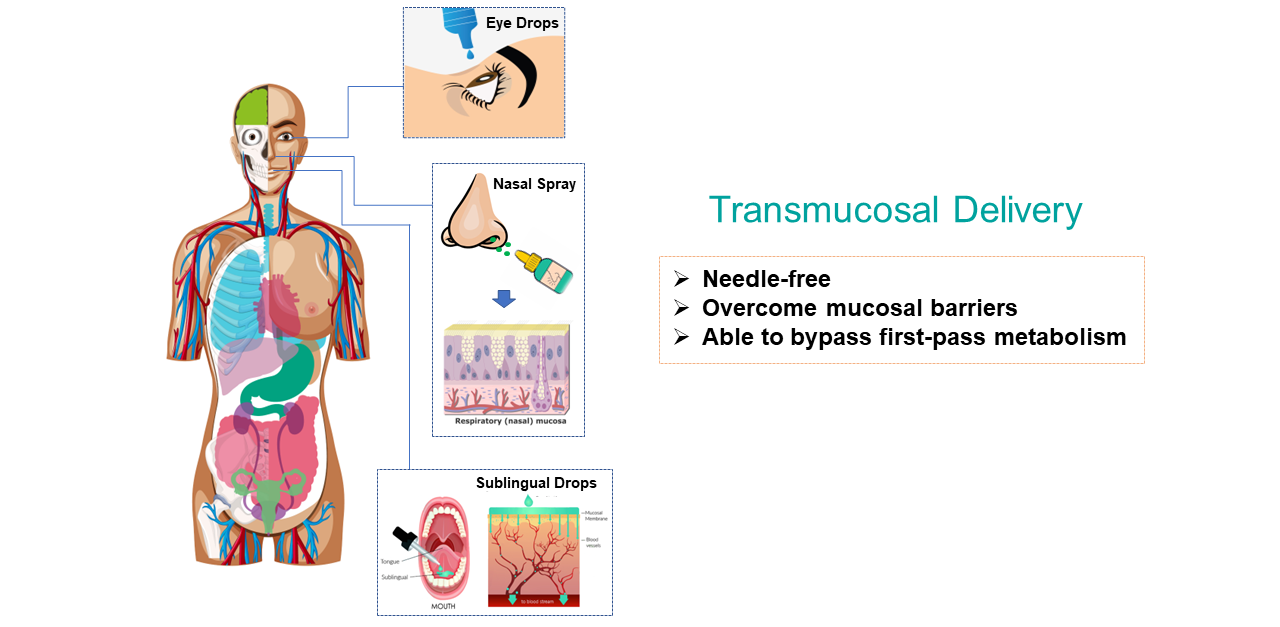
Protein drugs often require needle-based delivery, such as subcutaneous, intramuscular, and intravenous injection. This causes significant discomfort to patients, especially those who suffer from chronic diseases and require frequent and long-term doses. We are developing technologies that overcome mucosal and epithelial barriers to enable systemic absorption or enhanced penetration of proteins from the mucosal sites, including nasal, sublingual, and eye. Protein drugs are simply mixed with these delivery vehicles and then applied to the mucosal sites via sprays or drops. Potential applications include delivery of insulin for diabetes and monoclonal antibodies for autoimmune diseases and cancers.
Gene Therapy: Nucleic Acid Delivery Using Lipid Nanoparticles
Shyh-Dar Li
Investigator
Summary
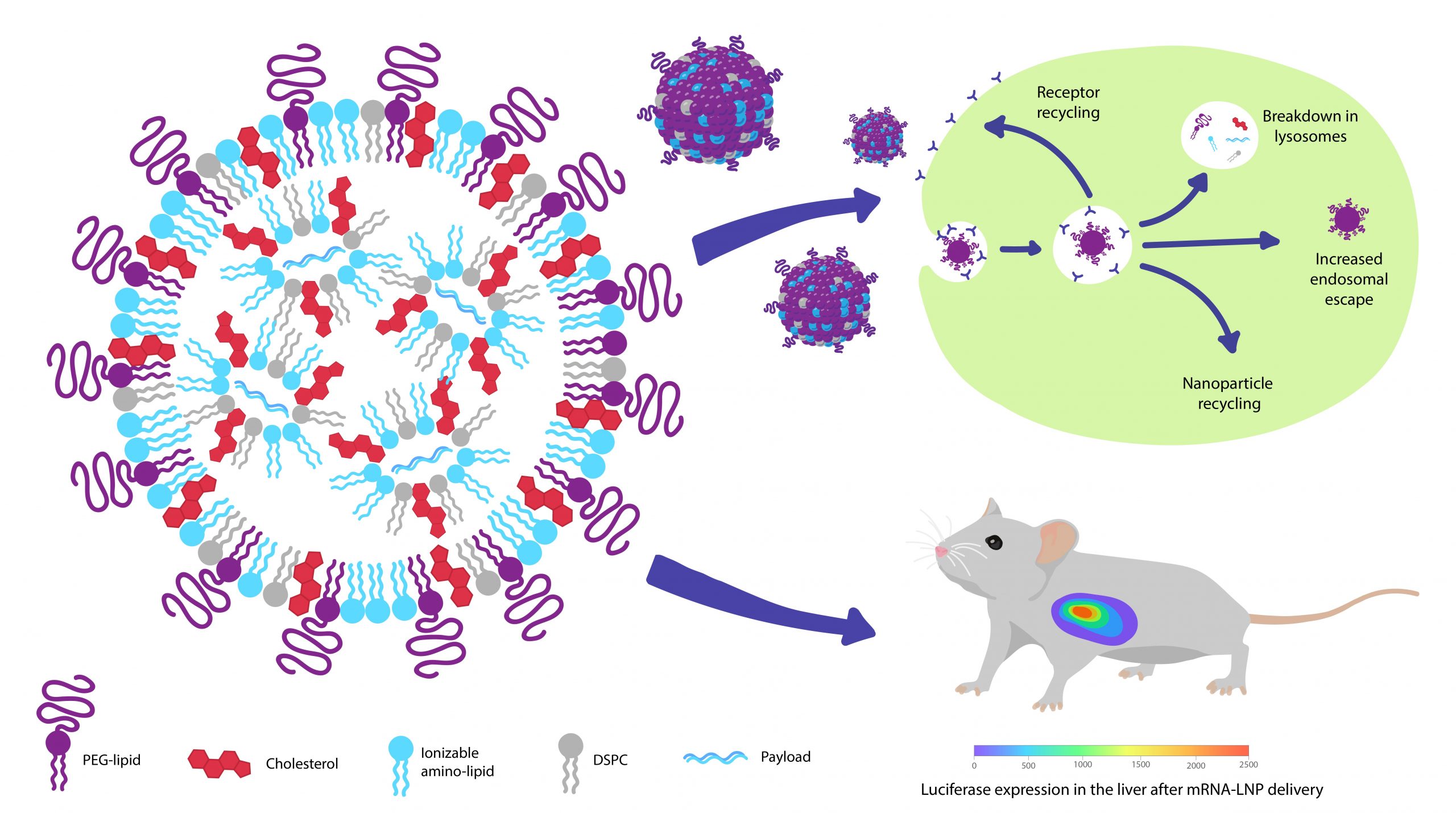
Lipid nanoparticle (LNP) is the state-of-the-art approach for nucleic acid delivery and gene therapy. However, only <2% of the nucleic acid delivered by the standard LNP formulation is released in the cytosol of cells for gene regulation. We are synthesizing novel lipids and polymers to replace the lipid components in the LNP formulation to increase gene delivery efficiency. This technology can be applied to vaccine development and gene therapy for genetic disorders.
Immunotherapy for Cancer
Shyh-Dar Li
Investigator
Summary
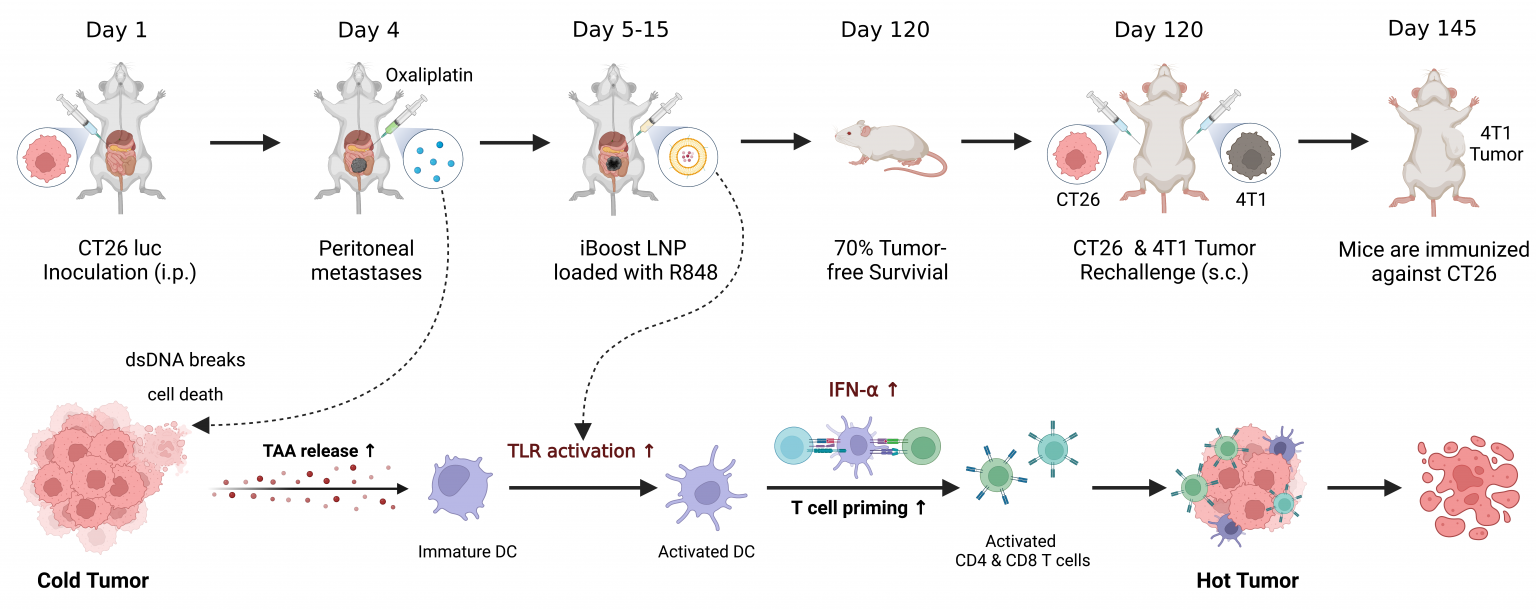
iBoost is a lipid nanoparticle formulation designed to retain payloads in the tumor microenvironment (TME). In a mouse model of peritoneal metastasis of colon cancer, iBoost largely focuses Resiquimod, a TLR agonist, to the antigen presenting cells (APCs) in the peritoneum while delaying its systemic absorption. As a result, iBoost increases the release of IFN-alpha in the TME. iBoost promotes T-cell priming to CD4+ and CD8+ T cells and potentiates the antitumor efficacy against this aggressive tumor model in combination with Oxaliplatin, a chemo drug. This new combination therapy is safe and effective in this model, curing 60–70% of the mice. Additionally, iBoost serves as an in-situ cancer vaccine. No cancer growth was observed in the cured mice post tumor re-challenge, suggesting the development of specific immunity, which provides great potential for cancer immunotherapy.
Developing new assays and inhibitors of Signal Transducer and Activator of Transcription (STAT) proteins
Brent Page
Investigator
Summary
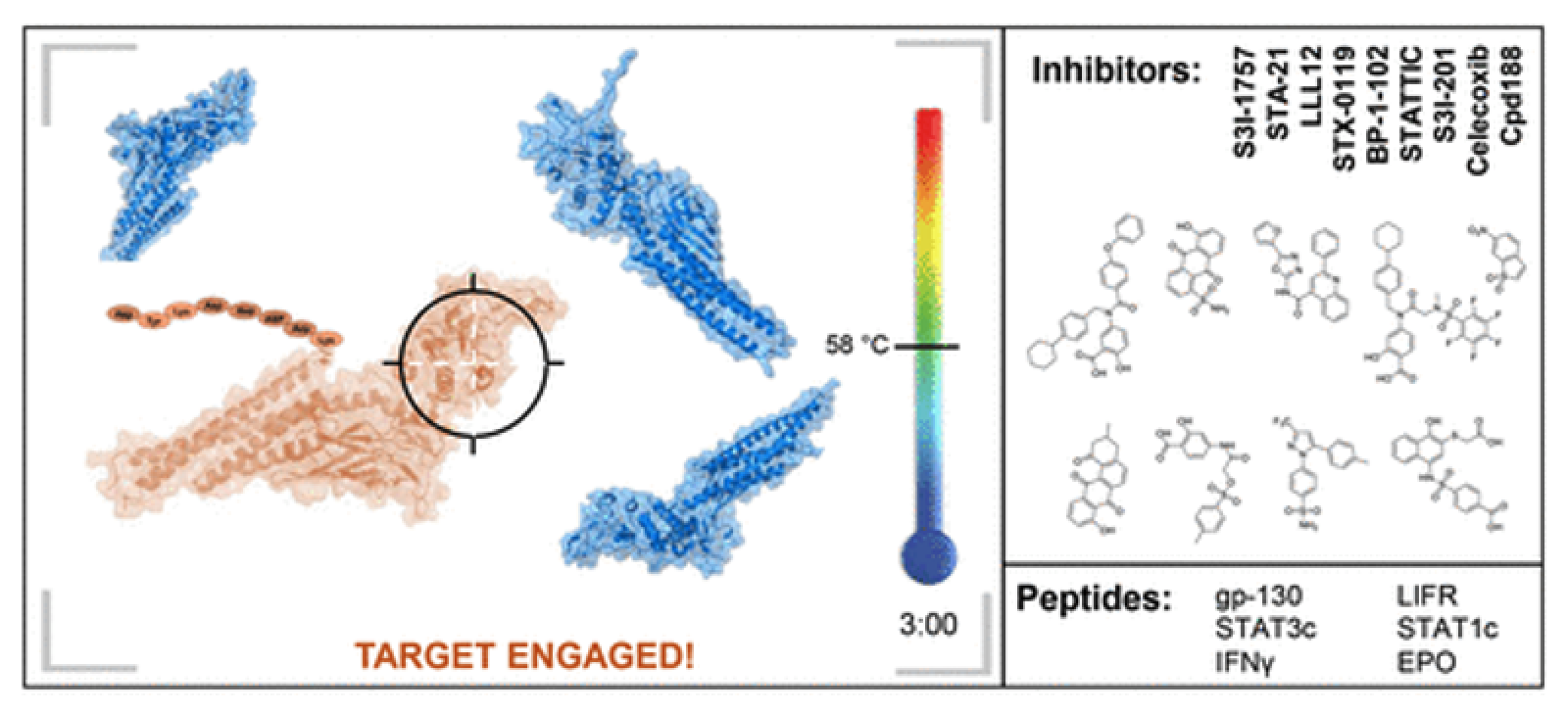
STAT proteins are important signaling molecules that mediate the cellular response to extracellular cytokine or growth factor stimulation. Deregulation of STAT signaling is commonly found in cancer and inflammatory conditions such as rheumatoid arthritis. While many STAT inhibitors have been reported, primarily focusing on STAT3 inhibitors as anti-cancer agents, our recently developed assays have shown that many of these compounds do not directly bind to STAT proteins in cells, and instead may bind to upstream regulators of STATs. We are now using these same assays to develop novel STAT1 and STAT3 inhibitors and are optimizing compounds that were identified from recent high-throughput screening campaigns.
Exploiting the anti-cancer activity of Pyrimethamine analogues
Brent Page, Adam Frankel
Investigators
Summary
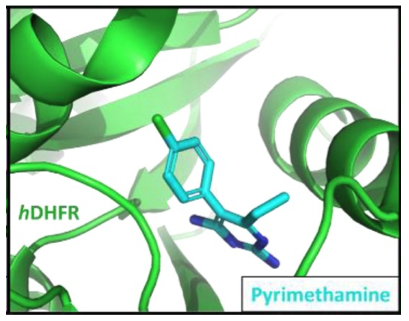
Pyrimethamine is a clinically approved anti-malaria drug that was recently discovered to inhibit STAT3 activity in cancer cells and induce promising anti-cancer activity. In the malaria parasite, pyrimethamine binds to dihydrofolate reductase (DHFR), however, this was not initially suspected to be the target for pyrimethamine in cancer cells. Recent work by our group has confirmed that DHFR is the primary target for pyrimethamine in human cells and we are working closely with the David Frank Laboratory at the Dana-Farber Cancer Institute to decipher the link between DHFR and STAT3 inhibition. Ongoing efforts in our lab aim to optimize the activity of new DHFR inhibitors that we hope further develop into promising anti-cancer agents.
Extracellular vesicles for liquid biopsy development in cancer
Karla Williams
Investigators
Summary
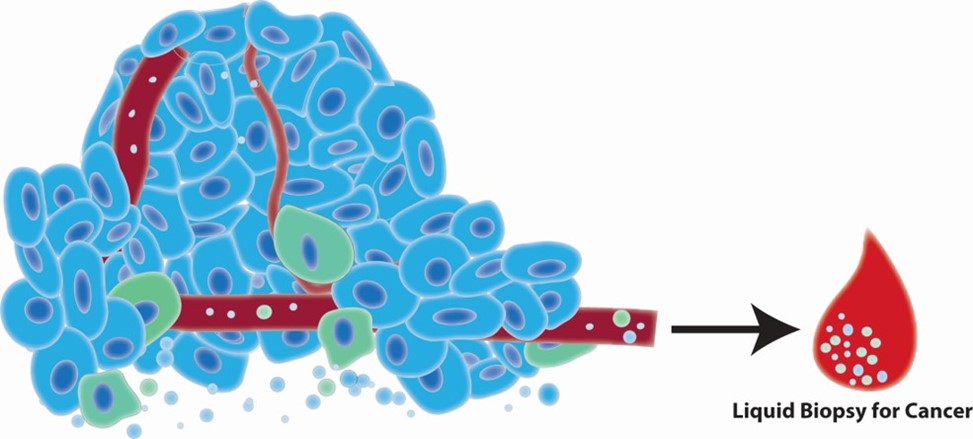
Developing biomarkers aimed to diagnosis and prognosis cancer is currently an intense area of research. Various approaches are being pursued using cells, circulating DNA/RNA, metabolites, protein, or extracellular vesicles (EVs), present in patient blood, urine, or other body fluids. EVs are nanosized, lipid-bilayer enclosed, particles which are rapidly shed from tumor cells and readily detectable in the blood. Given that the EV cargo is representative of the cell of origin, EVs hold great promise as a liquid biopsy for cancer. This suggests that strategies aimed at profiling the biomarker composition of EVs using bodily fluids have the potential to report an individual’s status of health versus cancer. We have explored the ability of EVs to report on breast cancer using targeted approaches, in combination with nanoscale flow cytometry, and using a proteomics approach to identify biomarkers of disease. Currently, we are working on a research project focused on identifying novel biomarkers of early-stage breast cancer. Here, we are focused on isolating these small particles from patient blood and interrogating their content. This work has the potential to discover biomarkers that could be used as a stand-alone screening platform, or in combination with current screening practices to help improve the detection of breast cancer at its earliest, most treatable, stage.
Translating the immune glyco-code
Simon Wisnovsky
Investigator
Summary
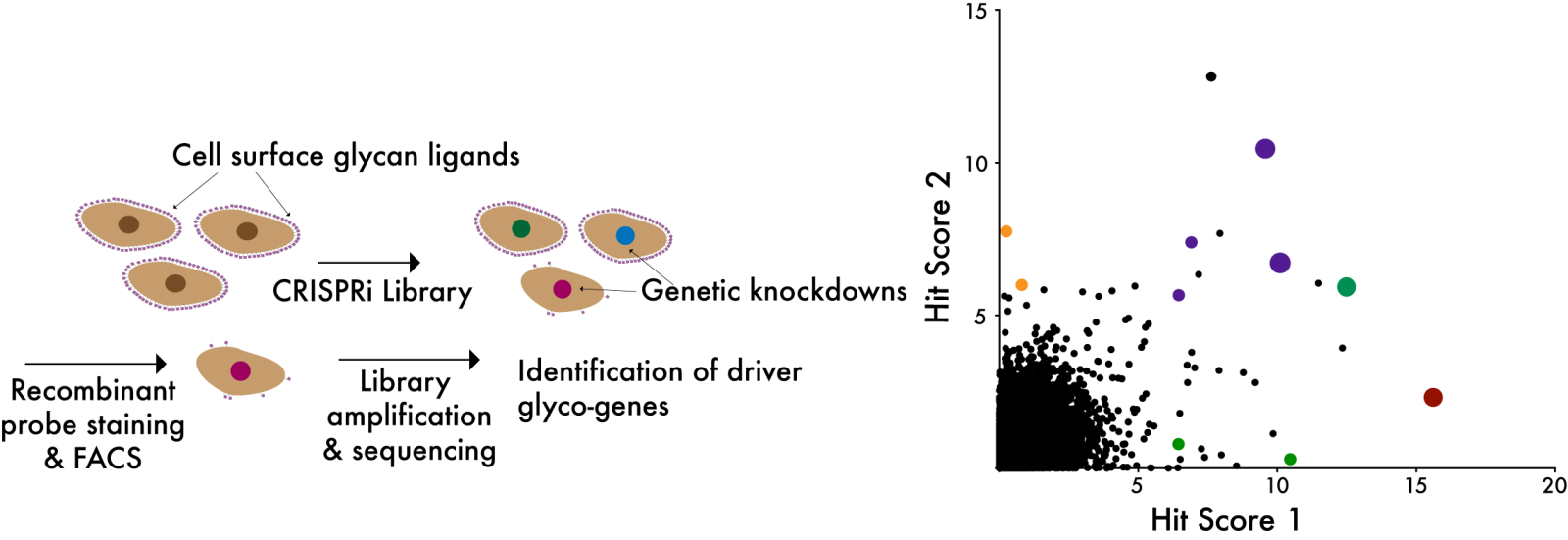
Prof. Wisnovsky's lab studies the cell surface glycome, a dense network of sugar molecules that coats the surface of every living cell. The glycome plays a fundamental role in regulating the activity of our immune system, helping immune cells to distinguish normal, healthy cells from abnormal cells and invading pathogens. In diseases like cancer, the structure of the cell-surface glycome becomes profoundly altered, allowing tumour cells to escape immune detection. Prof. Wisnovsky's group applies cutting-edge CRISPR genomic screening technologies to better understand the complex genetic mechanisms that regulate these changes in cellular glycosylation. The overarching goal of his research is to identify druggable pathways that can be targeted to modulate the cell-surface glycome, generating new therapeutic options for the treatment of cancer and autoimmune disease.
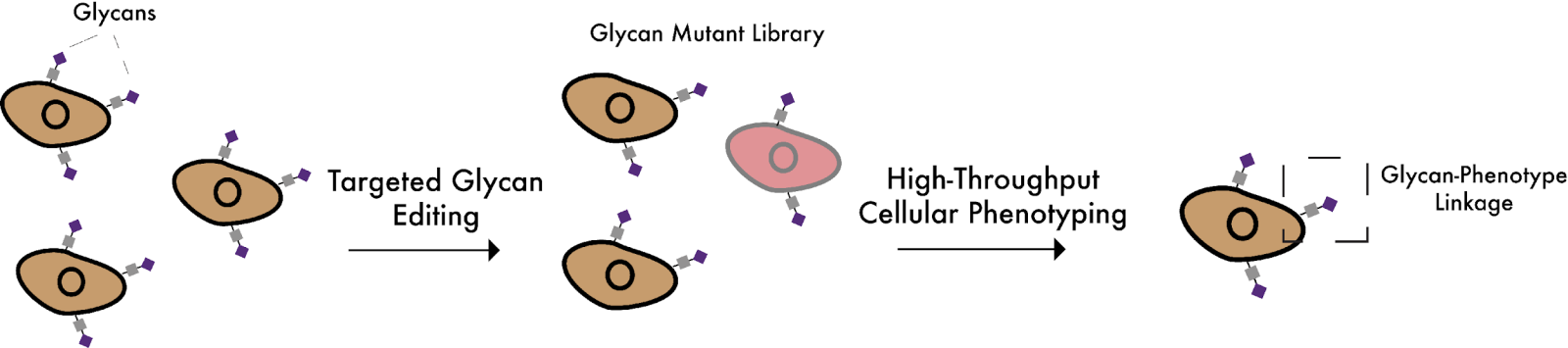
New technologies for glycome-wide screening
Hundreds of distinct carbohydrate structures (glycans) are attached to thousands of different protein scaffolds on the cell surface. The Wisnovsky lab is building new CRISPR-based technologies that will allow us to directly screen the function of these glycans in a pooled, high-throughput manner. We hope these tools will create new opportunities in translational glycoscience.